数据集
宫颈组织病理图像数据集-LDCH
The LDCH dataset contains 917 cervical histopathological whole-slide images: Cervicitis (156), CINI (305), CINII (296), CINIII (162), Cancer (25). Among them, 286 images were released with local annotations to mark out several lesion areas. The areas can be classified into 9 categories: CINI, CINII, CINIII, CINII involve gland, CINIII involve gland, gland, gland squamous metaplasia, koilocyte and squamous epithelium.
The images were collected by Landing Artificial Intelligence Center for Pathological Diagnosis. They scanned the histopathological sections at 40x magnification with the scanner produced by Jiangfeng. For each image, there were at least two deputy chief pathologists to scrutinize it for labelling.
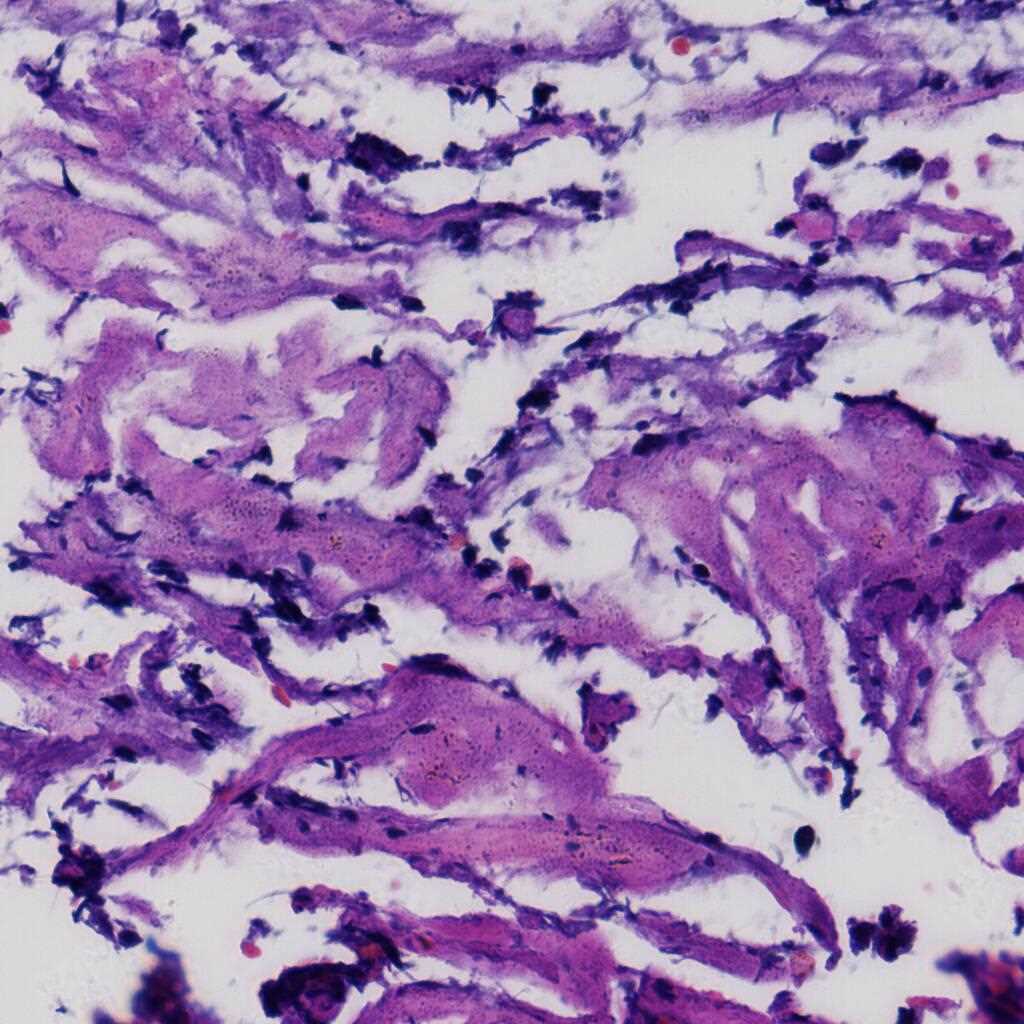
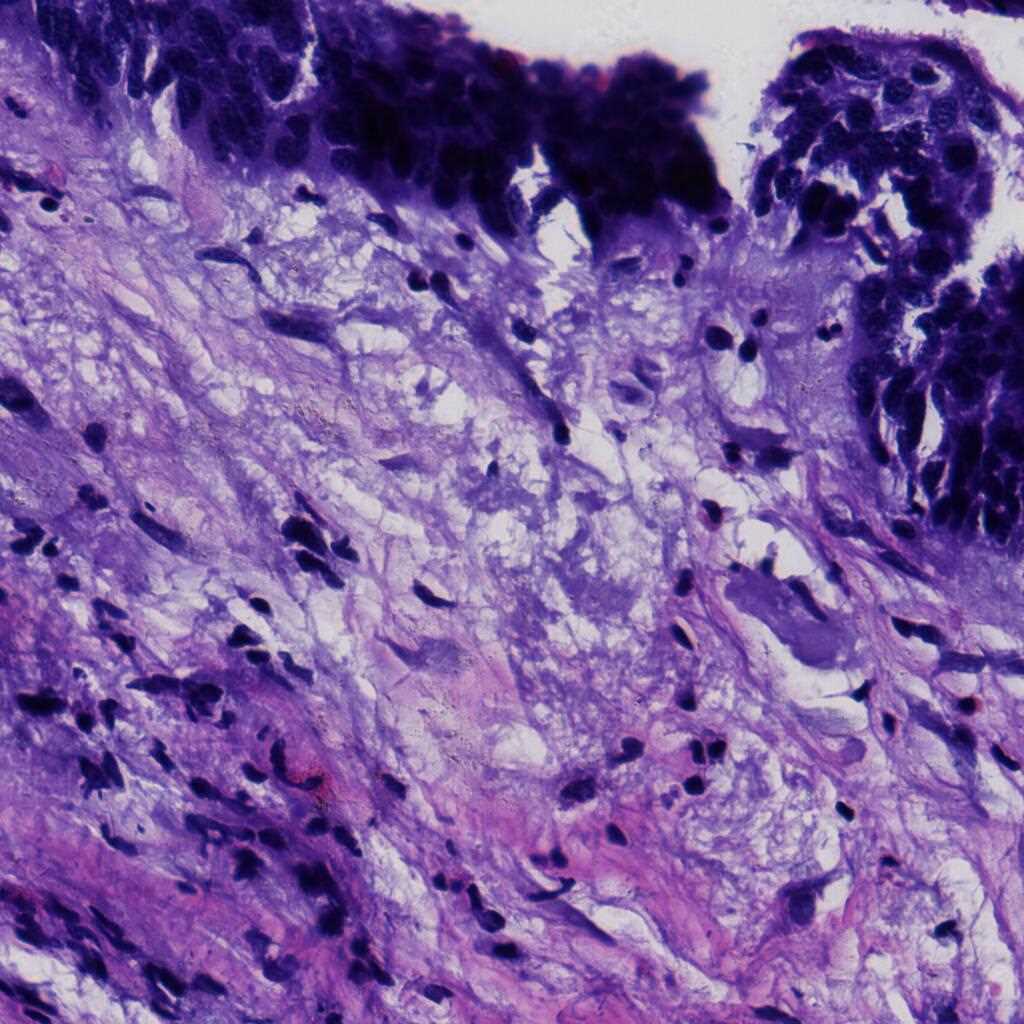
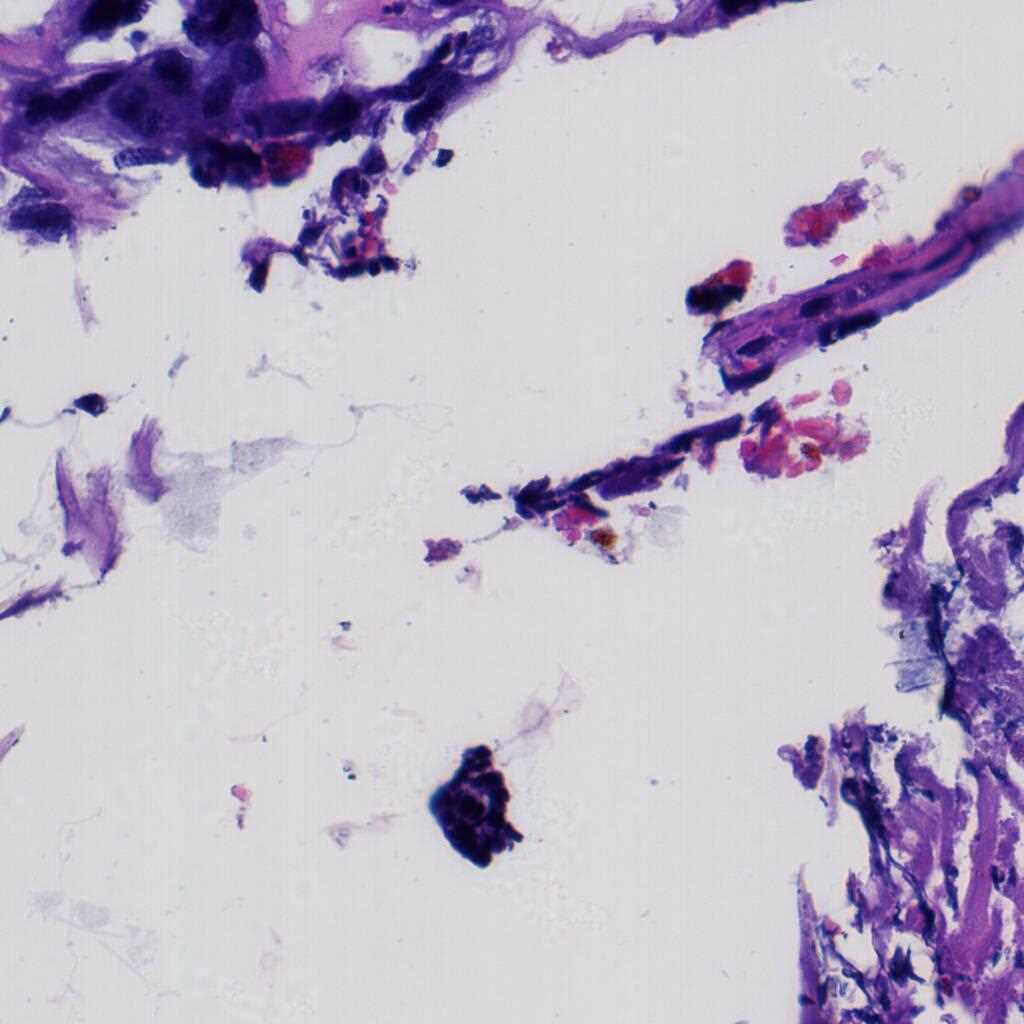
Using the LDCH dataset, please cite this:
[1] Peng Jiang, Juan Liu*, Lang Wang, Jing Feng, Dehua Cao, Baochuan Pang. Classifying Cervical Histopathological Whole Slide Images via Deep Multi-Instance Transfer Learning. 2022 IEEE International Conference on Bioinformatics and Biomedicine (BIBM), Las Vegas, NV, USA, 2022, pp. 2302-2307. DOI: 10.1109/BIBM55620.2022.9995014. [论文下载/paper download] [数据集下载/dataset download]
外周血白细胞图像数据集-LDWBC
The LDWBC dataset contains 22,645 images: Basophil (224), Monocyte (968), Eosinophil (539), Neutrophil (10,469) and Lymphocyte (10,445). The resolution of each image is 1280×1280 pixels. These images are collected from the samples from 150 microscope slides of 150 normal subjects. These samples are stained by Wright&Gimsa and then automatically scanned by a a light microscope with OLYMPUS BX41 and OLYMPUS Plan N 100x/1.25 to obtain corresponding digital images.
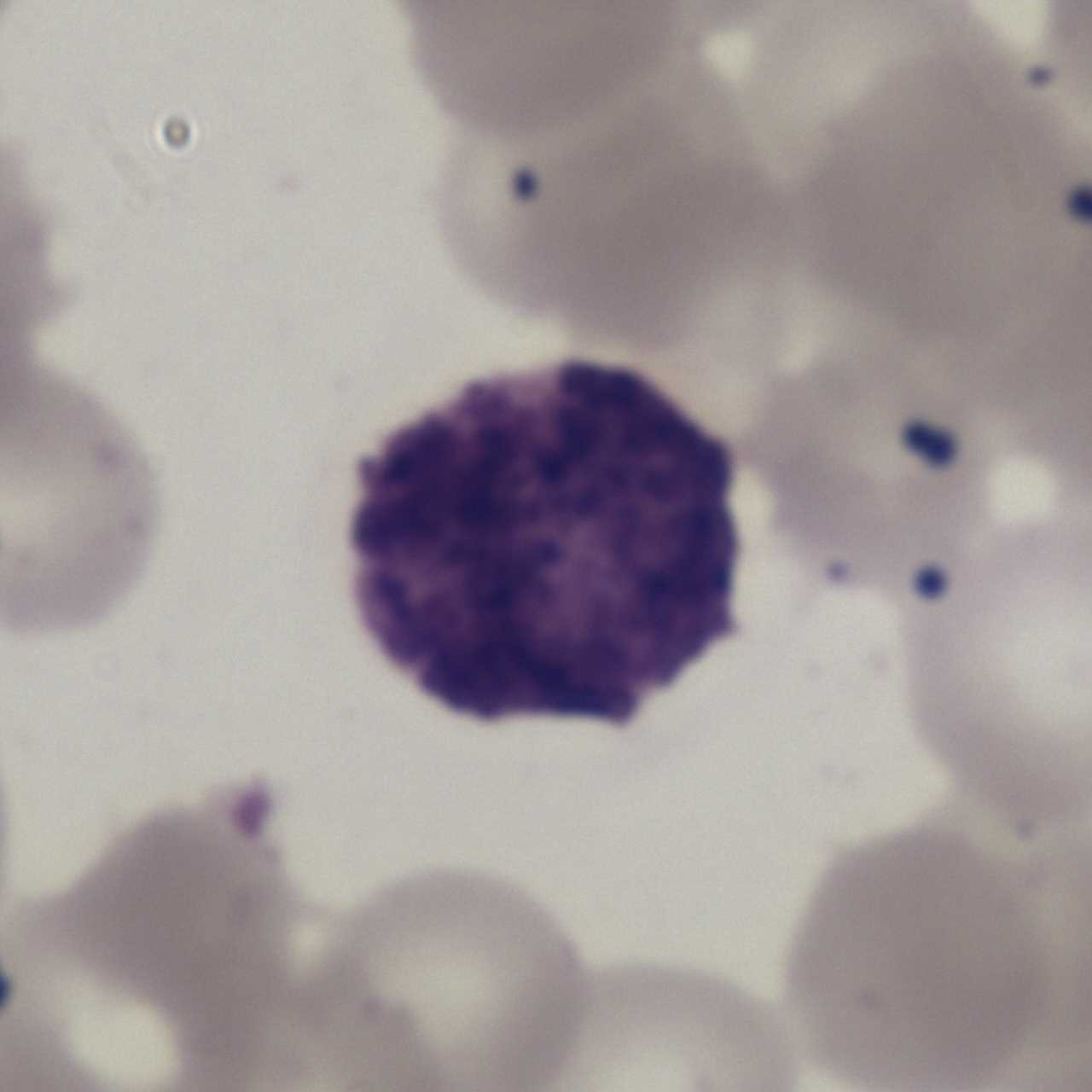
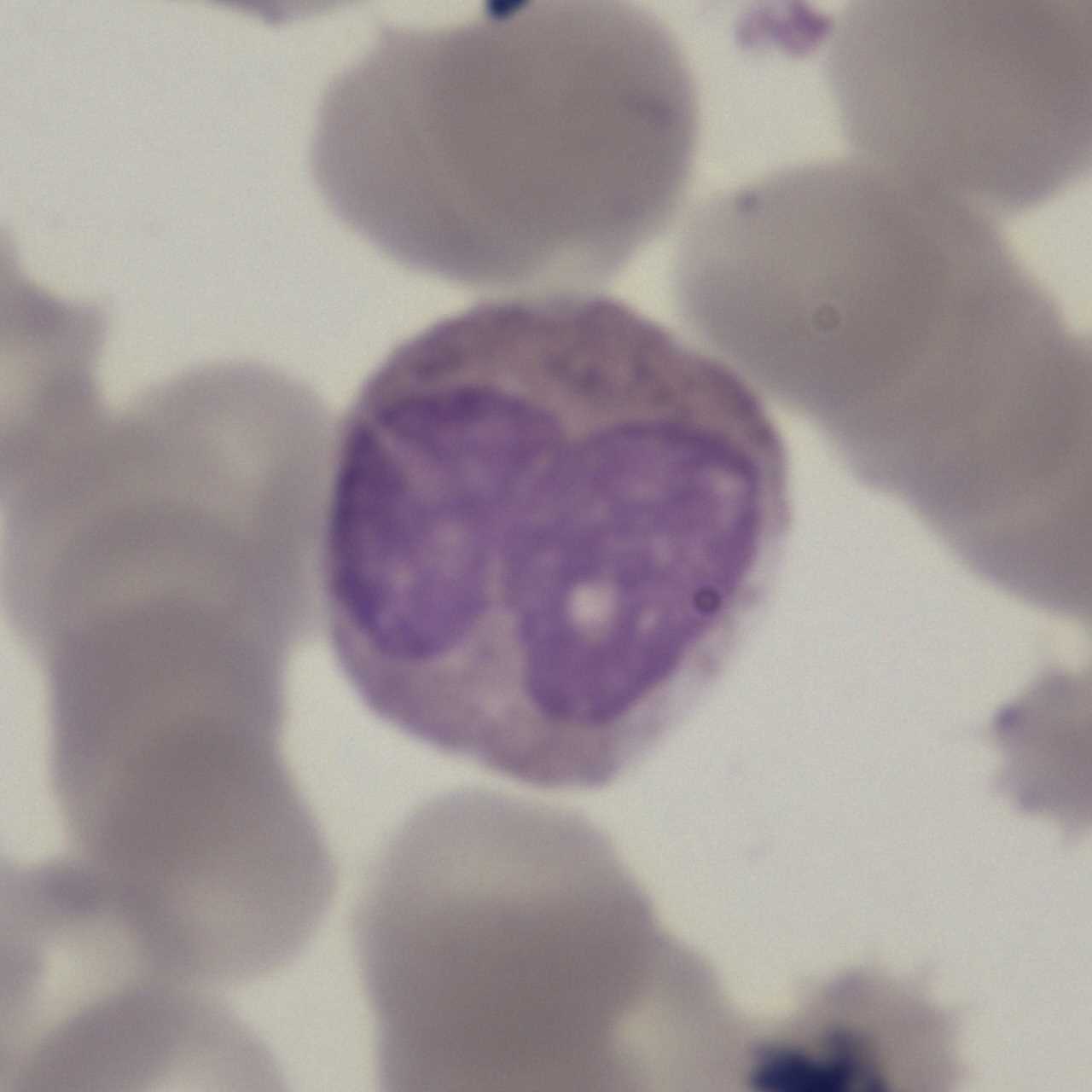

Using the LDWBC dataset, please cite this:
[1] Hua Chen, Juan Liu*, Chunbing Hua, Zhiqun Zuo, Jing Feng, Baochuan Pang, and Di Xiao. TransMixNet: An Attenion Based Double-Branch Model for White Blood Cell Classification and Its Training with the Fuzzified Training Data. 2021 IEEE International Conference on Bioinformatics and Biomedicine (BIBM2021), pp. 842-847. 2021. Doi: 10.1109/BIBM52615.2021.9669587 [论文下载/paper download] [数据集下载/dataset download]
宫颈细胞图像数据集1-LDCC
The LDCC dataset contains 198,952 images : Positive (60,238), Negative (25,001) and Junk (113,713), which is divided into training set and test set. The resolution of each image is 128×128 pixels. These images are collected from cervical cell samples from 2,312 participants. These samples are stained by hematoxylin and eosin (H&E) and then automatically scanned by a micro-scanning device with high resolution to obtain corresponding digital images.

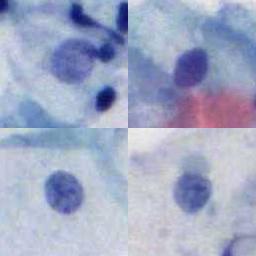

Using the LDCC dataset, please cite this:
[1] Hua Chen, Juan Liu*, Qing-Man Wen, Zhi-Qun Zuo, Jia-Sheng Liu, Jing Feng, Bao-Chuan Pang, Di Xiao. CytoBrain: Cervical Cancer Screening System Based on Deep Learning Technology[J]. Journal of Computer Science and Technology, 2021, 36(2): 347-360. Doi.org/10.1007/s11390-021-0849-3 [论文下载/paper download] [数据集下载/dataset download]
宫颈细胞图像数据集2-DSCC
The DSCC dataset contains 15,509 images: NORMAL (5,533), ASC (3,682) and SIL (6,294). Every cell in DSCC is manually labeled by professional cytologists. And, for each cell, 78 dimensional manual features are also provided. The resolution of each image is 128×128 pixels. These images are collected from cervical cell samples from 244 participants. These samples are stained by hematoxylin and eosin (H&E) and then automatically scanned by a micro-scanning device with high resolution to obtain corresponding digital images.
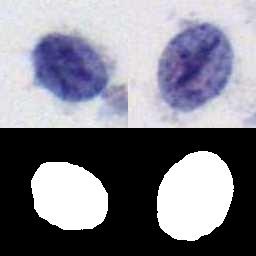
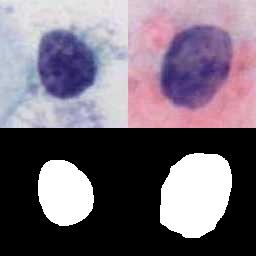

Using the DSCC dataset, please cite this:
[1] Hua Chen, Juan Liu*, Yu Jin, Baochuan Pang, Dehua Cao and Di Xiao. DSCC: A Data Set of Cervical Cell Images for Cervical Cytology Screening. International Journal of Data Mining and Bioinformatics, 2023, 27(1-3). DOI: 10.1504/ijdmb.2022.130325 [论文下载/paper download] [数据集下载/dataset download]
宫颈细胞图像数据集3-LTCCD
The LTCCD dataset contains 10,294 images : normal (3,350), ASCUS(1,744), ASC-H (1,600), LSIL (2,150) and HSIL (1,450). The resolution of each image is 128×128 pixels. These images are collected from cervical cell samples from 238 specimens. All specimens were produced into liquid-based preparations using Thinprep Cytological with the H&E staining method. All specimens were automatically scanned by a digital slide scanner (LD-Cyto-1004 adapted from Olympus BX43 microscope) in 20× objective lenses.
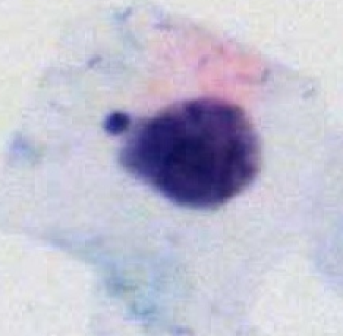
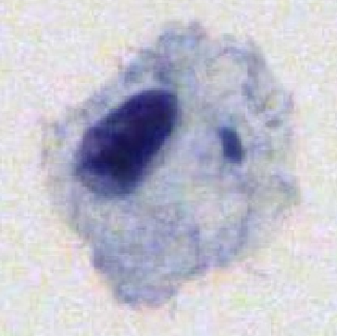

Using the LTCCD dataset, please cite this:
[1] Peng Jiang, Juan Liu*, Hua Chen, Cheng Li, Baochuan Pang, Dehua Cao. Channel Spatial Collaborative Attention Network for Fine-grained Classification of Cervical Cells. Neural Information Processing. ICONIP 2022. Communications in Computer and Information Science, vol 1793. Springer, Singapore. Doi.org/10.1007/978-981-99-1645-0_45 [论文下载/paper download] [数据集下载/dataset download]

